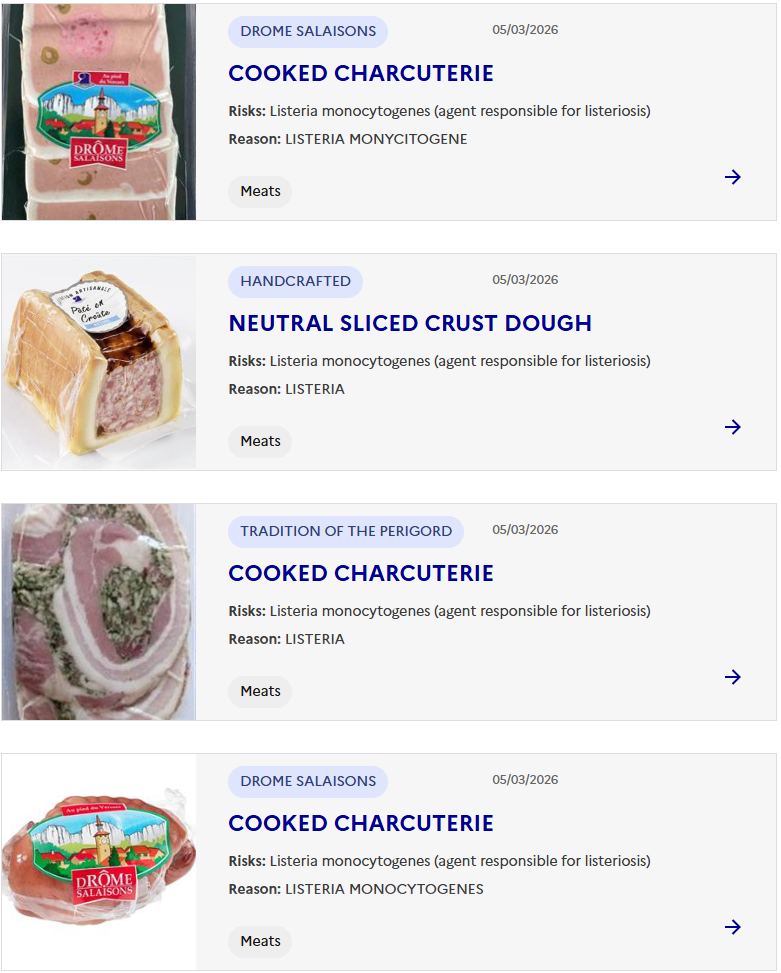
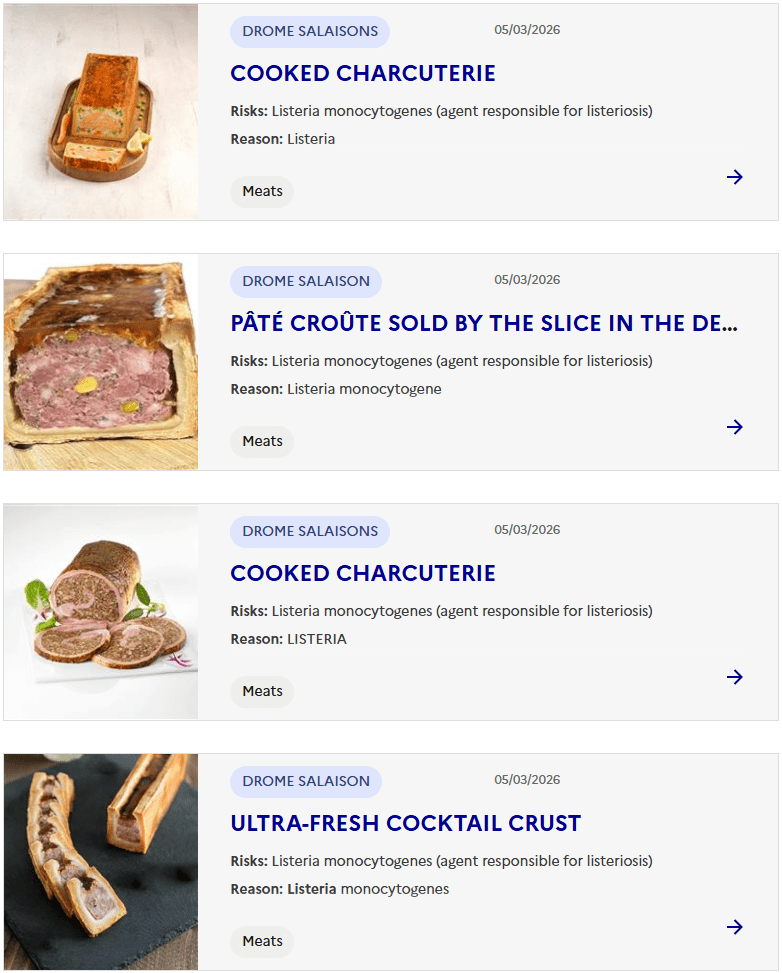
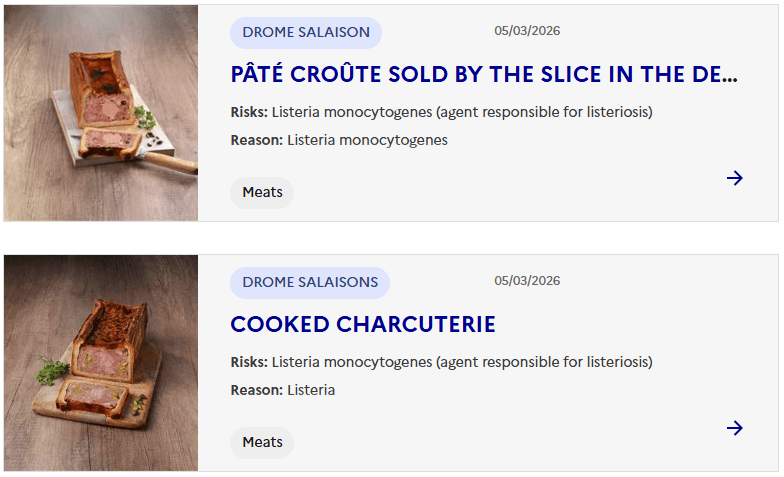
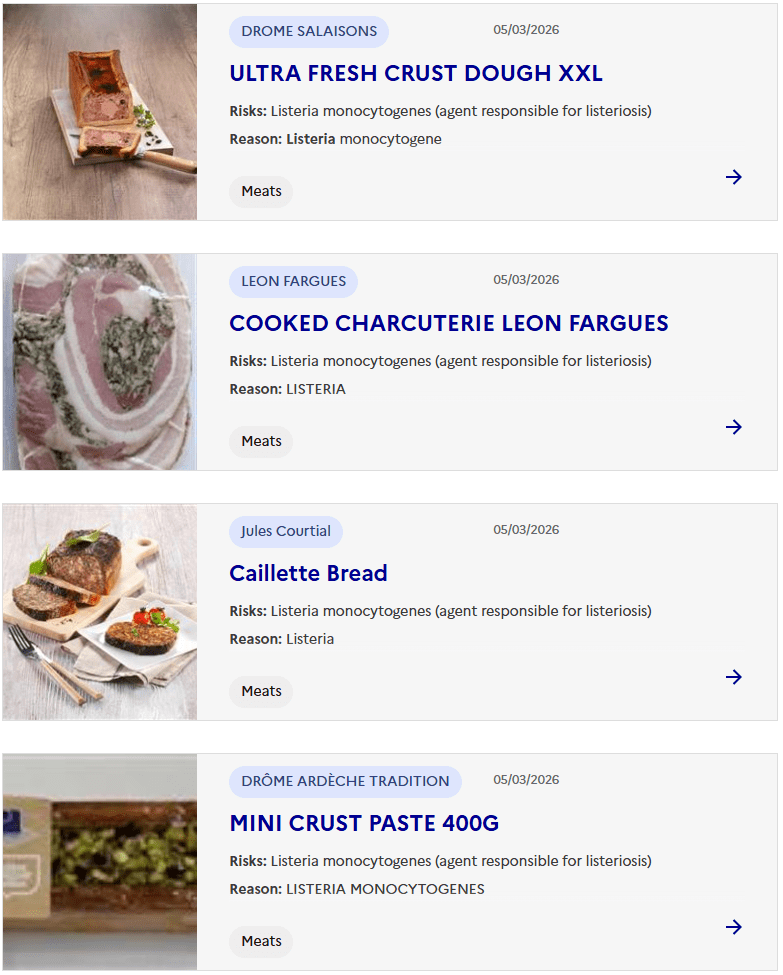
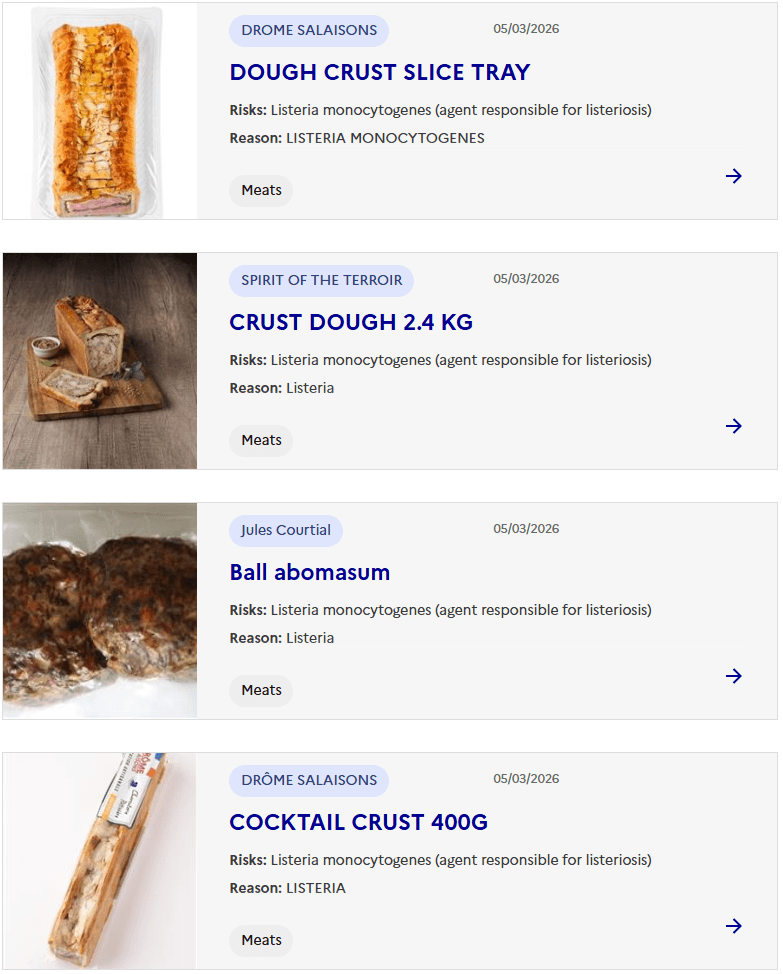
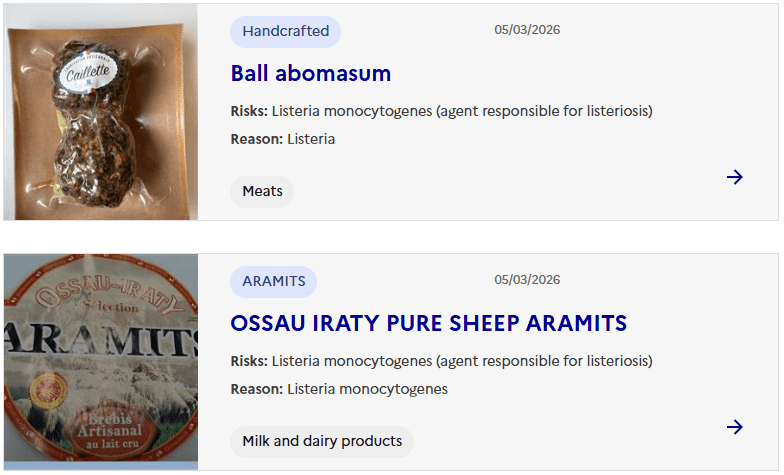
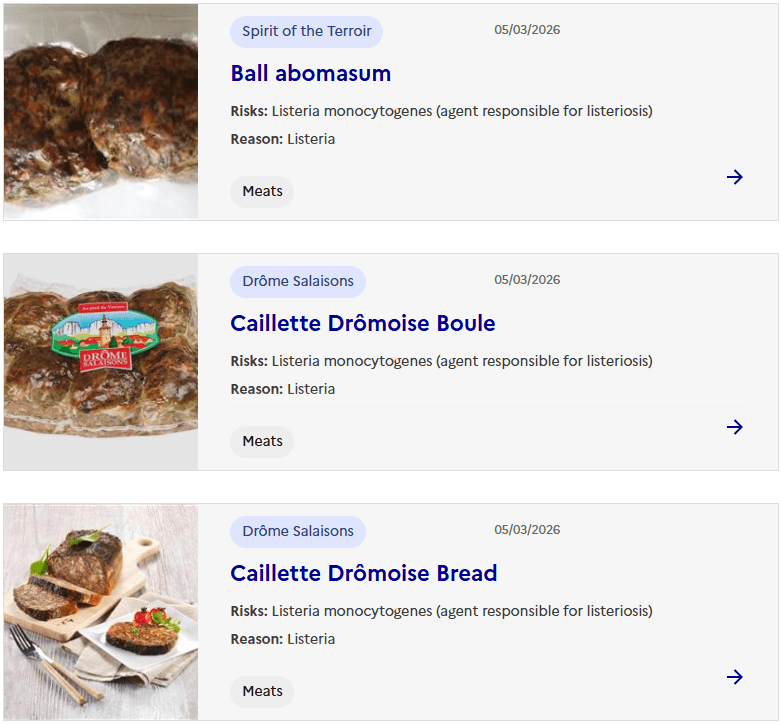
Sacor recalls Bastides Saucisson Sec because of contamination with Salmonella
Salmonella
Share this:
- Share on Facebook (Opens in new window) Facebook
- Email a link to a friend (Opens in new window) Email
- Share on X (Opens in new window) X
- Share on Pinterest (Opens in new window) Pinterest
- Share on LinkedIn (Opens in new window) LinkedIn
- Share on Reddit (Opens in new window) Reddit
- Share on WhatsApp (Opens in new window) WhatsApp
- Share on Mastodon (Opens in new window) Mastodon
- Share on Bluesky (Opens in new window) Bluesky
- More