Archives
-
Join 346 other subscribers
KSWFoodWorld
Blog Stats
- 438,557 Views
Category Archives: WGS
Research – Whole-Genome Sequencing of Shiga Toxin-Producing Escherichia coli for Characterization and Outbreak Investigation
Shiga toxin-producing Escherichia coli (STEC) causes high frequencies of foodborne infections worldwide and has been linked to numerous outbreaks each year. Pulsed-field gel electrophoresis (PFGE) has been the gold standard for surveillance until the recent transition to whole-genome sequencing (WGS). To further understand the genetic diversity and relatedness of outbreak isolates, a retrospective analysis of 510 clinical STEC isolates was conducted. Among the 34 STEC serogroups represented, most (59.6%) belonged to the predominant six non-O157 serogroups. Core genome single nucleotide polymorphism (SNP) analysis differentiated clusters of isolates with similar PFGE patterns and multilocus sequence types (STs). One serogroup O26 outbreak strain and another non-typeable (NT) strain, for instance, were identical by PFGE and clustered together by MLST; however, both were distantly related in the SNP analysis. In contrast, six outbreak-associated serogroup O5 strains clustered with five ST-175 serogroup O5 isolates, which were not part of the same outbreak as determined by PFGE. The use of high-quality SNP analyses enhanced the discrimination of these O5 outbreak strains into a single cluster. In all, this study demonstrates how public health laboratories can more rapidly use WGS and phylogenetics to identify related strains during outbreak investigations while simultaneously uncovering important genetic attributes that can inform treatment practices.
Posted in Decontamination Microbial, Food Micro Blog, Food Microbiology, Food Microbiology Blog, Food Microbiology Research, Food Microbiology Testing, microbial contamination, Microbial growth, Microbiological Risk Assessment, Microbiology, Microbiology Investigations, Microbiology Risk, MLST, PFGE, Shigatoxin, STEC, STEC E.coli, WGS
Research – Large Multicountry Outbreak of Invasive Listeriosis by a Listeria monocytogenes ST394 Clone Linked to Smoked Rainbow Trout, 2020 to 2021
Whole-genome sequencing (WGS) has revolutionized surveillance of infectious diseases. Disease outbreaks can now be detected with high precision, and correct attribution of infection sources has been improved. Listeriosis, caused by the bacterium Listeria monocytogenes, is a foodborne disease with a high case fatality rate and a large proportion of outbreak-related cases. Timely recognition of listeriosis outbreaks and precise allocation of food sources are important to prevent further infections and to promote public health. We report the WGS-based identification of a large multinational listeriosis outbreak with 55 cases that affected Germany, Austria, Denmark, and Switzerland during 2020 and 2021. Clinical isolates formed a highly clonal cluster (called Ny9) based on core genome multilocus sequence typing (cgMLST). Routine and ad hoc investigations of food samples identified L. monocytogenes isolates from smoked rainbow trout filets from a Danish producer grouping with the Ny9 cluster. Patient interviews confirmed consumption of rainbow trout as the most likely infection source. The Ny9 cluster was caused by a MLST sequence type (ST) ST394 clone belonging to molecular serogroup IIa, forming a distinct clade within molecular serogroup IIa strains. Analysis of the Ny9 genome revealed clpY, dgcB, and recQ inactivating mutations, but phenotypic characterization of several virulence-associated traits of a representative Ny9 isolate showed that the outbreak strain had the same pathogenic potential as other serogroup IIa strains. Our report demonstrates that international food trade can cause multicountry outbreaks that necessitate cross-border outbreak collaboration. It also corroborates the relevance of ready-to-eat smoked fish products as causes for listeriosis.
IMPORTANCE Listeriosis is a severe infectious disease in humans and characterized by an exceptionally high case fatality rate. The disease is transmitted through consumption of food contaminated by the bacterium Listeria monocytogenes. Outbreaks of listeriosis often occur but can be recognized and stopped through implementation of whole-genome sequencing-based pathogen surveillance systems. We here describe the detection and management of a large listeriosis outbreak in Germany and three neighboring countries. This outbreak was caused by rainbow trout filet, which was contaminated by a L. monocytogenes clone belonging to sequence type ST394. This work further expands our knowledge on the genetic diversity and transmission routes of an important foodborne pathogen.
Posted in Decontamination Microbial, food bourne outbreak, Food Illness, Food Micro Blog, Food Microbiology, Food Microbiology Blog, Food Microbiology Testing, Foodborne Illness, foodborne outbreak, foodbourne outbreak, Illness, Listeria, Listeria monocytogenes, Listeria Smoked Fish, listeriosis, microbial contamination, Microbial growth, Microbiological Risk Assessment, Microbiology, Microbiology Investigations, Microbiology Risk, outbreak, WGS
Research – Multi-country outbreak of Salmonella Virchow ST16 infections linked to the consumption of meat products containing chicken meat
Abstract
Since June 2017, a persistent cross-border outbreak of Salmonella Virchow ST16 has been ongoing in five European Union/European Economic Area (EU/EEA) countries, the United Kingdom (UK), and the United States (US). A total of 210 cases have been reported from the following countries: Denmark (2), France (111), Germany (26), Ireland (4), the Netherlands (34), the UK (32), and the US (1). Among the interviewed cases (55), hospitalisation rates ranged from 16.7% (2/12) in the UK, to 29.4% (5/17) and 38.5% (10/26) in France and Germany, respectively. No deaths have been reported. A majority of cases have been linked to local restaurants serving kebab meat. The number of confirmed cases represents only a small proportion of all infections in the EU/EEA, partly due to the varying sequencing capacities of countries.
The comparison of the representative outbreak strains with the available genome profiles of S. Virchow ST16 from non-human isolates, revealed that most of the matching isolates belonged to broiler meat and broiler-related environments, thereby supporting the hypothesis of chicken meat as a vehicle of infections.
The available information from case interviews, traceback investigations, and whole genome sequencing (WGS) cluster analysis, showed that kebab meat products containing contaminated chicken meat are the likely vehicles of infections, and that the clone has been circulating in the EU poultry meat production chain at least in France, Germany, and the Netherlands. In the absence of batch numbers of the contaminated kebab products and related Salmonella testing information, the source(s) of the infections could not be established.
New infections are likely to occur in the EU/EEA affecting any age group, until further investigations are performed to identify the source(s) and point(s) of contamination along the chicken meat production chain, including the primary production upstream lines. This will allow appropriate control measures to be implemented.
Posted in Decontamination Microbial, food bourne outbreak, Food Illness, Food Micro Blog, Food Microbiology, Food Microbiology Blog, Food Microbiology Research, Food Microbiology Testing, Foodborne Illness, foodborne outbreak, foodbourne outbreak, Illness, microbial contamination, Microbial growth, Microbiological Risk Assessment, Microbiology, Microbiology Investigations, Microbiology Risk, outbreak, Salmonella, Salmonella in Chicken, WGS
Research – Whole-Genome Sequence Comparisons of Listeria monocytogenes Isolated from Meat and Fish Reveal High Inter- and Intra-Sample Diversity
Interpretation of whole-genome sequencing (WGS) data for foodborne outbreak investigations is complex, as the genetic diversity within processing plants and transmission events need to be considered. In this study, we analyzed 92 food-associated Listeria monocytogenes isolates by WGS-based methods. We aimed to examine the genetic diversity within meat and fish production chains and to assess the applicability of suggested thresholds for clustering of potentially related isolates. Therefore, meat-associated isolates originating from the same samples or processing plants as well as fish-associated isolates were analyzed as distinct sets. In silico serogrouping, multilocus sequence typing (MLST), core genome MLST (cgMLST), and pangenome analysis were combined with screenings for prophages and genetic traits. Isolates of the same subtypes (cgMLST types (CTs) or MLST sequence types (STs)) were additionally compared by SNP calling. This revealed the occurrence of more than one CT within all three investigated plants and within two samples. Analysis of the fish set resulted in predominant assignment of isolates from pangasius catfish and salmon to ST2 and ST121, respectively, potentially indicating persistence within the respective production chains. The approach not only allowed the detection of distinct subtypes but also the determination of differences between closely related isolates, which need to be considered when interpreting WGS data for surveillance.
Posted in food contamination, food handler, Food Hazard, Food Hygiene, Food Inspections, Food Micro Blog, Food Microbiology, Food Microbiology Blog, Food Microbiology Research, Food Microbiology Testing, Food Pathogen, food recall, Food Safety, Food Safety Alert, Food Safety Management, Food Testing, Listeria, Listeria monocytogenes, MLST, Research, WGS
USA – Outbreak Investigation of Salmonella: Seafood (October 2022)
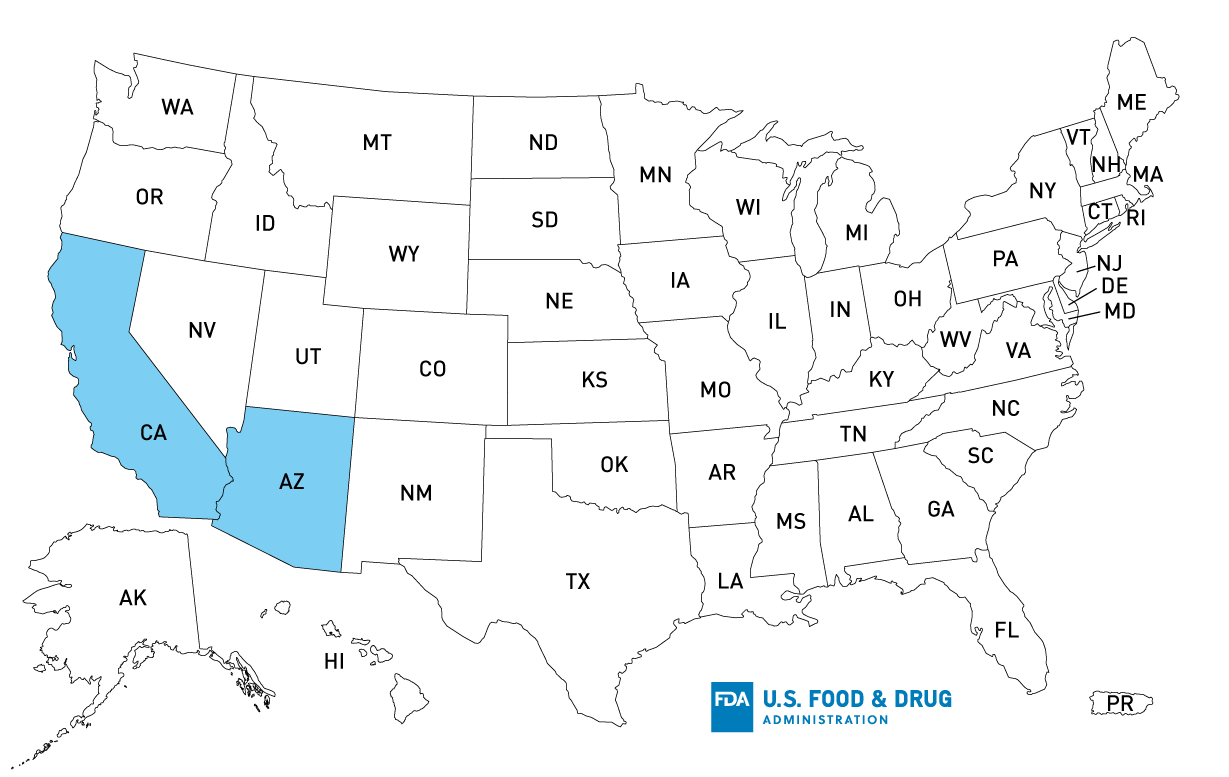
The FDA, along with CDC and state and local partners, is investigating a multistate outbreak of Salmonella Litchfield infections linked to fresh, raw salmon supplied to restaurants in California and Arizona by Mariscos Bahia, Inc.
Based on epidemiologic information provided by CDC and interviews conducted by state and local public health officials, of 16 people interviewed, 12 reported eating sushi, sashimi, or poke. Of those interviewed, 11 people remembered details about the type of fish consumed and 9 report eating raw salmon before getting sick. The FDA’s investigation traced the distribution of fresh, raw salmon back to Mariscos Bahia, Inc.
In addition, the FDA collected an environmental sample that included multiple swabs at Mariscos Bahia, Inc. (Pico Rivera, CA). Multiple environmental swabs collected at the facility are positive for Salmonella and subsequent Whole Genome Sequencing (WGS) analysis is ongoing. The WGS completed to date indicates the Salmonella detected in at least one of the swabs from the facility matches the outbreak strain. While epidemiological evidence indicates that ill people consumed fresh, raw salmon processed at this firm, the presence of Salmonella in the processing environment indicates that additional types of fish processed in the same area of the facility could also be contaminated which includes fresh, raw halibut, Chilean seabass, tuna, and swordfish. Salmon, halibut, Chilean seabass, tuna, and swordfish processed in Marisco Bahia Inc.’s Pico Rivera, CA, facility could have also been sent to the Mariscos Bahia, Inc. facilities in Phoenix, AZ and then sent to restaurants.
The firm is cooperating with the FDA investigation and has agreed to initiate a voluntary recall. As a part of the firm’s voluntary recall, the firm will contact its direct customers who received recalled product.
The FDA’s investigation is ongoing. Updates to this advisory will be provided as they become available.
Recommendation
According to Mariscos Bahia, Inc., seafood was only sold directly to restaurants in California and Arizona and would not be available for purchase by consumers in stores.
Restaurants should check with their suppliers and not sell or serve salmon, halibut, Chilean seabass, tuna, and swordfish received fresh, not frozen from Mariscos Bahia, Inc. (Pico Rivera, CA and Phoenix, AZ) on or after June 14, 2022. If restaurants received these fish and then froze it, they should not sell or serve it. Restaurants should also be sure to wash and sanitize locations where these fish from Mariscos Bahia, Inc. were stored or prepared.
Consumers eating salmon, halibut, Chilean seabass, tuna, and swordfish at a restaurant in California or Arizona should ask whether the fish is from Mariscos Bahia, Inc and was received fresh, not frozen.
Map of U.S. Distribution
Case Count Map Provided by CDC
Case Counts
Total Illnesses: 33
Hospitalizations: 13
Deaths: 0
Last illness onset: September 18, 2022
States with Cases: AZ (11), CA (21), IL (1)
Product Distribution*: AZ, CA
*Distribution has been confirmed for states listed, but product could have been distributed further, reaching additional states
Posted in CDC, Decontamination Microbial, food bourne outbreak, Food Illness, Food Micro Blog, Food Microbiology, Food Microbiology Blog, Food Microbiology Research, Food Microbiology Testing, Foodborne Illness, foodborne outbreak, foodbourne outbreak, Illness, microbial contamination, Microbial growth, Microbiological Risk Assessment, Microbiology, Microbiology Investigations, Microbiology Risk, outbreak, Salmonella, WGS
Research – Phenotypic and genotypic characterization of salmonella Enteritidis isolated from two consecutive Food-Poisoning outbreaks in Sichuan, China
Abstract
Salmonella enterica serotype Enteritidis (SE) is a primary pathogen that causes foodborne diseases in humans. Although whole-genome sequencing (WGS) -based typing analyses have been increasingly used to investigate food-poisoning outbreaks, they are rarely applied to the epidemiology of multiple Salmonella outbreaks in Sichuan, China. This study therefore isolated SE from patients and food of two consecutive food-poisoning outbreaks during 2020 in Sichuan, China. We tracked outbreak origin using epidemiological investigation, serotyping, antimicrobial susceptibility testing (AST), pulsed-field gel electrophoresis (PFGE), and WGS. We also determined phylogenetic relationships using PFGE, whole and core genome multilocus sequence typing (wg/cgMLST), and whole-genome single nucleotide polymorphism (wgSNP) analyses. Epidemiological investigations identified a correlation between cake consumption and food poisoning. Thirteen strains isolated from patients and one strain isolated from the cake were confirmed as SE. Among the 14 strains, only six shared the same AST pattern (AMP-AMS-Sul-STR). Isolates from patients and cakes were indistinguishable in PFGE results. All four methods, namely PFGE, wgMLST, cgMLST, and wgSNP were appropriate for bacterial typing in SE-related outbreak investigation. However, wgSNP can assign 12 SE strains from the first outbreak to one cluster and assign two SE strains from the second outbreak to another cluster, while PFGE, wgMLST, cgMLST did not successfully distinguish the SE strains from different outbreaks. Thus, we conclude that SNP-based phylogenetic analysis might be a viable method for differentiating SE strains at the outbreak level.
Posted in food bourne outbreak, Food Micro Blog, Food Microbiology, Food Microbiology Blog, Food Microbiology Research, Food Microbiology Testing, foodborne outbreak, foodbourne outbreak, microbial contamination, Microbiological Risk Assessment, Microbiology, Microbiology Investigations, Microbiology Risk, MLST, outbreak, PFGE, Research, WGS
Research – Whole-Genome Analysis of Staphylococcus aureus Isolates from Ready-to-Eat Food in Russia
Abstract
This study provides a thorough investigation of a diverse set of antimicrobial resistant (AMR) Staphylococcus aureus isolates collected from a broad range of ready-to-eat (RTE) food in various geographic regions of Russia ranging from Pskov to Kamchatka. Thirty-five isolates were characterized using the whole genome sequencing (WGS) analysis in terms of clonal structure, the presence of resistance and virulence determinants, as well as plasmid replicon sequences and CRISPR/Cas systems. To the best of our knowledge, this is the first WGS-based surveillance of Russian RTE food-associated S. aureus isolates. The isolates belonged to fifteen different multilocus sequence typing (MLST)-based types with a predominant being the ones of clonal complex (CC) 22. The isolates studied can pose a threat to public health since about 40% of the isolates carried at least one enterotoxin gene, and 70% of methicillin-resistant (MRSA) isolates carried a tsst1 gene encoding a toxin that may cause severe acute disease. In addition, plasmid analysis revealed some important characteristics, e.g., Rep5 and Rep20 plasmid replicons were a “signature” of MRSA CC22. By analyzing the isolates belonging to the same/single strain based on cgMLST analysis, we were able to identify the differences in their accessory genomes marking their dynamics and plasticity. This data is very important since S. aureus isolates studied and RTE food, in general, represent an important route of transmission and dissemination of multiple pathogenic determinants. We believe that the results obtained will facilitate performing epidemiological surveillance and developing protection measures against this important pathogen in community settings. View Full-Text
Posted in Decontamination Microbial, Food Micro Blog, Food Microbiology, Food Microbiology Blog, Food Microbiology Research, Food Microbiology Testing, microbial contamination, Microbiological Risk Assessment, Microbiology, Microbiology Investigations, Microbiology Risk, MRSA, Research, Staphylococcus aureus, WGS
USA – FDA Issues Country-Wide Import Alert for Enoki Mushrooms from the Republic of Korea – Listeria monocytogenes
The FDA announced today that its Import Divisions may detain without physical examination, importations of enoki mushrooms from the ROK. This country-wide import alert, IA #25-21, “Detention Without Physical Examination of Enoki Mushrooms from Korea (the Republic of) due to Listeria monocytogenes,” was issued to protect public health and help prevent the importation of enoki mushrooms that could be linked to human infections. The FDA issues import alerts to help prevent potentially violative products from being distributed in the United States.
In fiscal year 2021, FDA testing revealed that 43% of enoki mushrooms sampled from the ROK were contaminated with Listeria monocytogenes (L. monocytogenes). L. monocytogenes is a human pathogen that can be found in moist environments, soil, water, decaying vegetation and animals, and can survive and even grow under refrigeration and other food preservation measures.
The sampling was conducted following an FDA investigation into a multistate outbreak spanning from 2016-2020. This outbreak linked multiple cases of human infections of L. monocytogenes to enoki mushrooms from the ROK. The outbreak accounted for a total of 36 U.S. cases reported across 17 states, 12 cases in Canada, and six cases in Australia. The 36 U.S. cases yielded 31 hospitalizations and four deaths.
From March 2020 through May 2022, state public health authorities conducted sampling of enoki mushrooms from U.S. retail locations. L. monocytogenes was detected in multiple state samples, which led to 21 recalls of enoki mushrooms in the United States. Nine of the recalls were linked to enoki mushrooms grown in the ROK and were confirmed by labelling, traceback, or whole genome sequencing (WGS).
After the 2020 outbreak, the FDA began implementing an Imported Specialty Mushroom Prevention Strategy, with a focus on enoki mushrooms, to protect public health and prevent future L. monocytogenes outbreaks in specialty imported mushrooms. The FDA’s prevention strategies are affirmative, deliberate approaches undertaken by the agency to limit or prevent the recurrence of a root cause that led to an outbreak or adverse incident.
Posted in food contamination, food handler, Food Hazard, Food Hygiene, Food Inspections, Food Micro Blog, Food Microbiology, Food Microbiology Blog, Food Microbiology Testing, Food Pathogen, food recall, Food Safety, Food Safety Alert, Food Safety Management, Food Testing, Listeria, Listeria monocytogenes, WGS
Research – Use of Whole Genome Sequencing by the Federal Interagency Collaboration for Genomics for Food and Feed Safety in the United States
This multiagency report developed by the Interagency Collaboration for Genomics for Food and Feed Safety provides an overview of the use of and transition to whole genome sequencing (WGS) technology for detection and characterization of pathogens transmitted commonly by food and for identification of their sources. We describe foodborne pathogen analysis, investigation, and harmonization efforts among the following federal agencies: National Institutes of Health; Department of Health and Human Services, Centers for Disease Control and Prevention (CDC) and U.S. Food and Drug Administration (FDA); and the U.S. Department of Agriculture, Food Safety and Inspection Service, Agricultural Research Service, and Animal and Plant Health Inspection Service. We describe single nucleotide polymorphism, core-genome, and whole genome multilocus sequence typing data analysis methods as used in the PulseNet (CDC) and GenomeTrakr (FDA) networks, underscoring the complementary nature of the results for linking genetically related foodborne pathogens during outbreak investigations while allowing flexibility to meet the specific needs of Interagency Collaboration partners. We highlight how we apply WGS to pathogen characterization (virulence and antimicrobial resistance profiles) and source attribution efforts and increase transparency by making the sequences and other data publicly available through the National Center for Biotechnology Information. We also highlight the impact of current trends in the use of culture-independent diagnostic tests for human diagnostic testing on analytical approaches related to food safety and what is next for the use of WGS in the area of food safety.
Research – New Challenges for Detection and Control of Foodborne Pathogens: From Tools to People
Contamination of foods by human pathogenic microorganisms is a major concern to both food safety and public health. The changes in consumers’ demand, the globalization of the food trade, and the progress on food production practices and processing technologies all pose new challenges for food industries and regulatory agencies to ensure the safety in food products.
With regard to microbiological safety, bacteria and viruses are the most common foodborne pathogens associated with both sporadic cases and outbreaks.
However, bacterial and viral microorganisms differ in terms of their behaviour in food matrices, their stability in food-related environments (e.g., food-contact surfaces, irrigating and processing waters), and their response to food processing technologies and controlling measures. Current methods do not meet all relevant criteria for effective monitoring plans, the main limitations being their sensitivity, the high workload and time requirement, and the inability to differentiate between viable and non-viable microorganisms. Thus, specific and sensitive methods need to be developed for their detection and quantification in com-plex matrices, such as food, for tracking their occurrence along the food chain to determine the sources of contamination, and for ultimately estimating the risk for consumers.
To fill these gaps, this Special Issue comprises four original research articles and are view paper focusing on the implementation of novel analytical techniques and approaches to foodborne pathogens along the food chain.
Zand and colleagues [1] reviewed the most recent advances of the application of flowcytometry (FCM) and fluorescence in situ hybridization (FISH) for the rapid detection and characterization of microbial contamination. FCM allows for a culture-independent quantification of microbial cells, also providing information on their physiological and structural characteristics which are relevant to assess their viability status. FISH is a nucleic acid-based method mainly applied in the medical and diagnostic fields. While FCM has been successfully used to detect and monitor microorganisms in water, state-of-the-art FCM and FISH protocols for food matrices still show significant limitations. The main pitfalls include complex sample preparation steps; the use of toxic substances; their limits of detection, especially for FISH assays; and the equipment price. Because of all these aspects, FCM and FISH have not yet gained considerable interest in food safety area for the detection of microbial pathogens. Future studies should focus on potential optimisation strategies for FCM and FISH protocols in food samples and their validation, as well as on the development of automated lab-on-chip solutions.
Moving to explore next-generation sequencing (NGS) applications in the produce industry, Truchado et al. [2] contributed to identify potential contamination niches of Listeria monocytogenes in a frozen vegetable processing plant. NGS is a sequencing technology that offers ultra-high throughput, a scalable and fast technique that allows the authors to characterize the isolates by a whole-genome sequencing (WGS) of 3multi locus sequence typing (MLST). The WGS analysis revealed the presence of four different sequence types (ST) contaminating 18% of the samples, including food contact surfaces (FCS), non-food contact surfaces (n-FCS), and final product. These ST were further classified into four different virulence types (VT) according to multi-virulence locus sequence typing (MVLST). Interestingly, an isolate detected in non-food-contact surfaces(n-FCS) also contaminated the final product, highlighting the relevant role of n-FCS as reservoir of L.monocytogenes that reached the final product.
Staphylococcus aureus is a foodborne pathogen considered to be one of the etiological agents of food-related disease outbreaks. Leng et al. [3] supported this Special Issue with a study on its control using the skin mucus extract of Channa argus as a source of antimicrobial compounds. Of interest, untargeted metabolomics were applied to decipherits antibacterial mechanism against S. aureus. Results indicated that the extract had great inhibitory action on its growth by inducing the tricarboxylic acid cycle and amino acid biosynthesis, which are the primary metabolic pathways that affect the normal physiological functions of biofilms.
The present collection includes a second contribution on the control of S. aureus authored by Kim and colleagues [4] who developed a real-time PCR method (qPCR) for the rapid detection and quantification of pathogenic Staphylococcus species.
Four specific molecular targets were identified based on pan-genome analysis, and results showed 100% specificity for 100 non-target reference strains with a detection limit as low as 102CFU/mL. Thus, the proposed method allows an accurate and rapid monitoring of Staphylococcus species and may help control staphylococcal contamination of food.
Moving to human viral pathogens, Macaluso et al. [5] reported the results of an investigation aimed to characterize the occurrence of human enteric viruses in shellfish, a food item with relevant risk for consumers. The study included data collected over two years on the prevalence of enteric virus contamination along the shellfish production and distribution chain in Sicily, Italy. The findings based on quantitative reverse transcription polymerase chain reactions (RT-qPCRs), as gold-standard molecular technique, showed that almost 6% of samples were contaminated with at least one enteric virus such as norovirus, hepatitis A virus, and/or hepatitis E virus. The origin of contaminated shellfish was traced back to Spain and several municipalities in Italy. Such contribution highlights the relevance
of routine monitoring programs to prevent foodborne transmission of enteric viruses and
preserve the health of consumers.
In summary, this Special Issue compile several contributions focused on novel technologies, approaches, and strategies demonstrated to be effective in controlling microbial contamination in food. All the articles provide valuable information to monitor and/or reduce contamination in food, food industry settings, and along the food chain. On a final note, the collection emphasizes the relevance of ensuring food safety and limiting the risk of microbiological contamination along the food chain to protect consumers.