Salmonella spp. are the third most common cause of bacterial food-borne illnesses worldwide and the second most commonly reported zoonotic agents in the European Union (EU). The bacterial genus Salmonella consists of Salmonella enterica and Salmonella bongori species. More than 2,500 serotypes of Salmonella enterica have been identified so far , many of them commonly infecting animals (e.g. poultry, pigs, cattle) and humans. The distribution of predominant serovars in each country are affected by changes in the global food and livestock trade, international travel, and human migration.
Salmonella enterica subsp. enterica serovar Bareilly (S. Bareilly) belongs to the C1 serogroup (antigenic formula 6, 7, 14: y: 1,5) and was first identified in India in 1928. In the United Kingdom (UK), 31% of all S. Bareilly human cases identified between 2005 and 2009 were attributed to travel from India. Since 2004, S. Bareilly has most commonly been isolated from spices. Contaminated mung bean seeds were the likely source of a S. Bareilly outbreak in the UK in 2010, with total of 231 cases. In an outbreak of salmonellosis in the United States, which comprised 410 cases of S. Bareilly across 28 states and the District of Columbia, tuna scrape imported from India was identified to be the source using whole genome sequencing (WGS)-based methods.
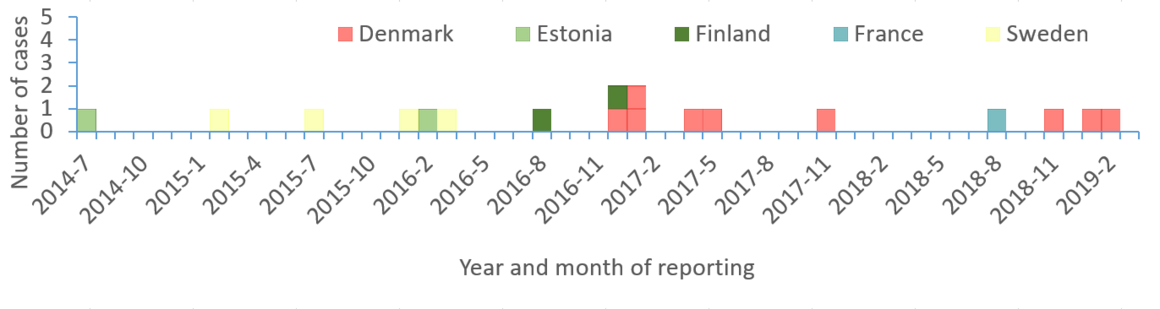
Since 2016, S. Bareilly has been among the top 20 Salmonella serotypes associated with human diseases in the European Union/European Economic Area (EU/EEA) . Between 2006 and 2016, S. Bareilly was among the top 25 serotypes detected in humans in the Czech Republic, with the annual incidence ranging from 0.04 to 0.23 per 100,000 inhabitants (data from the Czech national electronic communicable diseases notification system). According to data from the Czech national control programme for Salmonella in poultry, S. Bareilly was identified in broiler flocks with a prevalence of up to 0.06%.
Salmonellosis has been a mandatory notifiable disease in both the Czech Republic and Slovakia since 1951. Regional public health officers notify case-based data to the national electronic communicable diseases notification system (EpiDat/ISIN in the Czech Republic and the Epidemic Intelligence Information System (EPIS) in Slovakia). Both systems record data on all cases that meet the definition of a confirmed case in accordance with the European Commission Implementing Decision 2119/98/EC. The information on Salmonella serovar, which is provided by routine microbiological laboratories handling human samples, is included in the reporting systems. These laboratories typically test for a limited spectrum of serovars only, and S. Bareilly is usually not included. The Czech and Slovak National Reference Laboratories (NRLs) (the Czech NRL is a part of the National Institute of Public Health in Prague, the Slovak NRL is part of the Public Health Authority in Bratislava) provide serotyping of less common serovars and confirm results from routine microbiological laboratories on request.
There are several options to confirm the relatedness of Salmonella isolates in laboratories. Macro-restriction analysis followed by pulsed-field gel electrophoresis (PFGE) is usually a suitable method for the detection and investigation of Salmonella outbreaks. However, in some cases, it does not provide sufficient discriminatory power to distinguish outbreak isolates. Therefore, WGS-based typing methods are now increasingly applied as molecular epidemiology tools to assist in outbreak investigations.