Archives
-
Join 343 other subscribers
KSWFoodWorld
Blog Stats
- 429,644 Views
Category Archives: microbial contamination
Research – Microbiological Quality of Polish Artisanal Varietal Honeys
On the basis of routine microbiological tests and selected physicochemical parameters, the quality and food safety of Polish varietal honeys were evaluated. The study included 21 honey samples from 5 varieties (multifloral, honeydew, rapeseed, buckwheat and linden), in which the moisture and extract content, water activity, pH and free acids were determined, and the colony count, the presumptive Bacillus spp., the total fungal count and the presence of anaerobic spore-forming bacilli were examined. More than half (52%, 11/21) of the analyzed honeys contained fewer microorganisms than 10 cfu/g, and in the remaining samples, their numbers ranged from 5 × 101 cfu/g to 4.5 × 102 cfu/g. In all the honeys, the number of presumptive Bacillus spp. in 1 g was less than 10 cfu. In 81% (17/21) of the samples, the total count of fungi in 1 g of honey was less than 10 cfu, and the most contaminated was buckwheat honey (3 samples). The anaerobic spore-forming bacteria was detected in 0.1 g only in one sample of buckwheat honey. The values of the physicochemical parameters did not exceed the accepted limits, which indicated that the honey environment was unfavourable for the development of the tested microbial profile.
Research – First Report of Food Poisoning Due to Staphylococcal Enterotoxin Type B in Döner Kebab (Italy)
Abstract
Staphylococcal food poisoning results from the consumption of food contaminated by staphylococcal enterotoxins. In July 2022, the Turin local health board was notified of a suspected foodborne outbreak involving six children who had consumed döner kebab purchased from a takeaway restaurant. The symptoms (vomiting and nausea) were observed 2–3 h later. A microbiological analysis of the food samples revealed high levels (1.5 × 107 CFU/g) of coagulase-positive staphylococci (CPS). The immunoassay detected a contamination with staphylococcal enterotoxins type B (SEB). The whole genome sequencing of isolates from the food matrix confirmed the staphylococcal enterotoxin genes encoding for type B, which was in line with the SEB detected in the food. This toxin is rarely reported in staphylococcal food poisoning, however, because there is no specific commercial method of detection. The involvement of enterotoxin type P (SEP) was not confirmed, though the corresponding gene (sep) was detected in the isolates. Nasal swabs from the restaurant food handlers tested positive for CPS, linking them to the likely source of the food contamination.
Posted in Bacterial Toxin, Decontamination Microbial, Food Micro Blog, Food Microbiology, Food Microbiology Blog, Food Microbiology Research, Food Microbiology Testing, microbial contamination, Microbial growth, Microbiological Risk Assessment, Microbiology, Microbiology Investigations, Microbiology Risk, Staphylococcal Toxin, Staphylococcus aureus
Research – Listeria monocytogenes Biofilms in Food-Associated Environments: A Persistent Enigma

Abstract
Listeria monocytogenes (LM) is a bacterial pathogen responsible for listeriosis, a foodborne illness associated with high rates of mortality (20–30%) and hospitalisation. It is particularly dangerous among vulnerable groups, such as newborns, pregnant women and the elderly. The persistence of this organism in food-associated environments for months to years has been linked to several devastating listeriosis outbreaks. It may also result in significant costs to food businesses and economies. Currently, the mechanisms that facilitate LM persistence are poorly understood. Unravelling the enigma of what drives listerial persistence will be critical for developing more targeted control and prevention strategies. One prevailing hypothesis is that persistent strains exhibit stronger biofilm production on abiotic surfaces in food-associated environments. This review aims to (i) provide a comprehensive overview of the research on the relationship between listerial persistence and biofilm formation from phenotypic and whole-genome sequencing (WGS) studies; (ii) to highlight the ongoing challenges in determining the role biofilm development plays in persistence, if any; and (iii) to propose future research directions for overcoming these challenges.
Posted in Biofilm, Decontamination Microbial, Food Micro Blog, Food Microbiology, Food Microbiology Blog, Food Microbiology Research, Food Microbiology Testing, Listeria, Listeria monocytogenes, microbial contamination, Microbial growth, Microbiological Risk Assessment, Microbiology, Microbiology Investigations, Microbiology Risk
Research – Studying bacteriophage therapy in chickens

Fowl typhoid (FT) is among the most significant poultry bacterial diseases worldwide, mostly affecting developing countries.1 The causal agent of FT is Salmonella enterica Gallinarum. Although S enterica Gallinarum is egg-transmitted and produces lesions in chicks and poults, there is a much greater tendency to spread among growing or mature flocks. Mortality in young birds is possible but tends to be higher in older birds.2
In a poster presentation at the 2023 World Anti-Microbial Resistance Congress in Philadelphia, Pennsylvania, researchers from Purdue University in West Lafayette, Indiana, and the University of Veterinary and Animal Sciences (UVAS) Lahore, Pakistan, stated that, “Antibiotics are often used to prevent or control fowl typhoid; however, such practices contribute to the continually global challenge of antibiotic resistance.”1
“In previous studies, we developed a polyphage prototype that significantly reduced Salmonella Gallinarum in experimentally challenged chickens. Here, we evaluated the pact of treatment with the polyphage prototype on microbial communities surrounding the targeted bacteria.”1
Posted in Bacteriophage, Decontamination Microbial, Food Micro Blog, Food Microbiology, Food Microbiology Blog, Food Microbiology Research, Food Microbiology Testing, microbial contamination, Microbial growth, Microbiological Risk Assessment, Microbiology, Microbiology Investigations, Microbiology Risk, Phage, Salmonella, Salmonella in Chicken
Research -Effect of Sodium Nitrite, Nisin and Lactic Acid on the Prevalence and Antibiotic Resistance Patterns of Listeria monocytogenes Naturally Present in Poultry
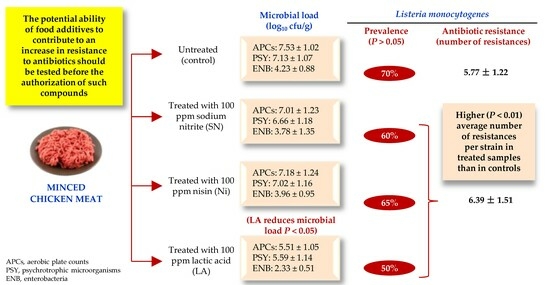
Abstract
The impact of treating minced chicken meat with sodium nitrite (SN, 100 ppm), nisin (Ni, 10 ppm) and lactic acid (LA, 3000 ppm) on the levels of some microbial groups indicating hygiene quality were investigated. Specifically, aerobic plate counts and culture-based counts of psychrotrophic microorganisms and enterobacteria were obtained. Additionally, the prevalence of Listeria monocytogenes and the resistance of 245 isolates from this bacterium to 15 antibiotics were documented. L. monocytogenes was isolated using the ISO 11290-1:2017 method and confirmed with polymerase chain reaction using the lmo1030 gene. Antibiotic resistance was established using the disc diffusion technique (EUCAST and CLSI criteria). Twenty-four hours after treatment, the microbial load (log10 cfu/g) was reduced (p < 0.05) relative to controls in those samples treated with LA, with counts of 5.51 ± 1.05 (LA-treated samples) vs. 7.53 ± 1.02 (control) for APC, 5.59 ± 1.14 (LA) vs. 7.13 ± 1.07 (control) for psychrotrophic microorganisms and 2.33 ± 0.51 (LA) vs. 4.23 ± 0.88 (control) for enterobacteria. L. monocytogenes was detected in 70% (control samples), 60% (samples receiving SN), 65% (Ni) and 50% (LA) (p > 0.05) of samples. All strains showed resistance to multiple antimicrobials (between 3 and 12). In all, 225 isolates (91.8%) showed a multi-drug resistant (MDR) phenotype, and one isolate (0.4%) showed an extensively drug-resistant (XDR) phenotype. The mean number of resistances per strain was lower (p < 0.01) in the control samples, at 5.77 ± 1.22, than in those receiving treatment, at 6.39 ± 1.51. It is suggested that the use of food additives might increase the prevalence of resistance to antibiotics in L. monocytogenes, although additional studies would be necessary to verify this finding by analyzing a higher number of samples and different foodstuffs and by increasing the number of antimicrobial compounds and concentrations to be tested.
Posted in Decontamination Microbial, Food Micro Blog, Food Microbiology, Food Microbiology Blog, Food Microbiology Research, Food Microbiology Testing, Listeria, Listeria monocytogenes, microbial contamination, Microbial growth, Microbiological Risk Assessment, Microbiology, Microbiology Investigations, Microbiology Risk
Research – Microbiological Quality and Safety of Fresh Quail Meat at the Retail Level
Abstract
The objective of this study was to evaluate the microbiological quality and safety of 37 fresh quail meats. Mesophiles, Pseudomonas spp., Enterobacteriaceae, and staphylococci counts were 5.25 ± 1.14, 3.92 ± 1.17, 3.09 ± 1.02, and 2.80 ± 0.64 log CFU/g, respectively. Listeria monocytogenes was detected in seven samples (18.92%). Campylobacter jejuni was detected in one sample (2.70%). Clostridium perfringens was not detected in any sample. The dominant bacteria were Pseudomonas spp. (30.46%), Micrococcaceae (19.87%), lactic acid bacteria (14.57%), and Enterobacteriaceae (11.92%). Brochotrix thermosphacta and enterococci were isolated to a lesser extent, 7.28% and 1.99%, respectively. The dominant Enterobacteriaceae found were Escherichia coli (42.53%). ESBL-producing E. coli was detected in one sample (2.70%), showing resistance to 16 antibiotics. Sixteen different Staphylococcus spp. and three Mammaliicoccus spp. were identified, the most common being S. cohnii (19.86%) and M. sciuri (17.02%). S. aureus and S. epidermidis were also found in one and four samples, respectively. Methicillin-resistant M. sciuri and S. warneri were found in 13.51% and 10.81% of quail samples, respectively. These bacteria showed an average of 6.20 and 18.50 resistances per strain, respectively. The high resistance observed in ESBL-producing E. coli and methicillin-resistant S. warneri is of special concern. Measures should be adopted to reduce the contamination of quail meat.
Posted in Brochothrix thermosphacta, Clostridium perfringens, Decontamination Microbial, Enterobacteriaceae, ESBL, Food Micro Blog, Food Microbiology, Food Microbiology Blog, Food Microbiology Research, Food Microbiology Testing, microbial contamination, Microbial growth, Microbiological Risk Assessment, Microbiology, Microbiology Investigations, Microbiology Risk, Pseudomonas, Staphylococcus aureus
Research – Natural preservatives for hams: Essential oil mixtures and major compounds’ efficacy against Clostridium sporogenes
Abstract
The demand for healthier products by consumers has prompted research into the use of essential oils (EOs), which are known for their antimicrobial properties. This study investigated the antimicrobial properties of EOs as a potential alternative to synthetic preservatives, specifically, cinnamon, clove, and oregano EOs, as well as their majority compounds (cinnamaldehyde, eugenol, and carvacrol) on Clostridium sporogenes inoculated in hams. The findings of the study revealed that cinnamon EO and its major compound cinnamaldehyde were the most efficient in inhibiting the growth of C. sporogenes, demonstrating the lowest minimum bactericidal concentration MBC (0.1%). The combination of oregano EO, cinnamon EO, cinnamaldehyde, and carvacrol led to a significant decrease in bacterial growth (approximately 3 log) after 28 days in hams. Furthermore, the presence of spores was not observed for 14 days (at 14°C) and 21 days (at 7°C) of storage, indicating a delay in sporulation. The treatments using the combination of EOs, and their major compounds had minimal impact on the color of the hams while maintaining the physicochemical characteristics of the product. This study demonstrates that EOs and their major compounds can be applied as natural preservatives in ham, offering a potential alternative for reducing the use of nitrites in various food types. The research emphasizes the antibacterial efficacy of cinnamon, clove, and oregano EOs, along with their major compounds, in inhibiting C. sporogenes in ham. The findings indicate that these natural alternatives could be valuable in preserving food products and reducing the reliance on synthetic preservatives.
Posted in Clostridium, Clostridium sprogenes, Decontamination Microbial, Food Micro Blog, Food Microbiology, Food Microbiology Blog, Food Microbiology Research, Food Microbiology Testing, microbial contamination, Microbial growth, Microbiological Risk Assessment, Microbiology, Microbiology Investigations, Microbiology Risk
Research – Viability and Virulence of Listeria monocytogenes in Poultry
Abstract
The prevalence of Listeria monocytogenes in 30 samples of poultry was determined using culture-dependent (isolation on OCLA and confirmation by conventional polymerase chain reaction -PCR-, OCLA&PCR) and culture-independent (real-time polymerase chain reaction, q-PCR) methods. L. monocytogenes was detected in 15 samples (50.0%) by OCLA&PCR and in 20 (66.7%) by q-PCR. The concentrations (log10 cfu/g) of L. monocytogenes (q-PCR) ranged from 2.40 to 5.22 (total cells) and from <2.15 to 3.93 (viable cells). The two methods, q-PCR using a viability marker (v-PCR) and OCLA&PCR (gold standard), were compared for their capacity to detect viable cells of L. monocytogenes, with the potential to cause human disease. The values for sensitivity, specificity and efficiency of the v-PCR were 100%, 66.7% and 83.3%, respectively. The agreement between the two methods (kappa coefficient) was 0.67. The presence of nine virulence genes (hlyA, actA, inlB, inlA, inlC, inlJ, prfA, plcA and iap) was studied in 45 L. monocytogenes isolates (three from each positive sample) using PCR. All the strains harbored between six and nine virulence genes. Fifteen isolates (33.3% of the total) did not show the potential to form biofilm on a polystyrene surface, as determined by a crystal violet assay. The remaining strains were classified as weak (23 isolates, 51.1% of the total), moderate (one isolate, 2.2%) or strong (six isolates, 13.3%) biofilm producers. The strains were tested for susceptibility to a panel of 15 antibiotics. An average of 5.11 ± 1.30 resistances per isolate was observed. When the values for resistance and for reduced susceptibility were taken jointly, this figure rose to 6.91 ± 1.59. There was a prevalence of resistance or reduced susceptibility of more than 50.0% for oxacillin, cefoxitin, cefotaxime, cefepime ciprofloxacin, enrofloxacin and nitrofurantoin. For the remaining antibiotics tested, the corresponding values ranged from 0.0% for chloramphenicol to 48.9% for rifampicin. The high prevalence and level of L. monocytogenes with numerous virulence factors in poultry underline how crucial it is to follow correct hygiene procedures during the processing of this foodstuff in order to reduce the risk of human listeriosis.
South African scientists sound warning after Listeria found in beef
South African researchers have warned about the risk of another outbreak after a study found Listeria in the beef sector.
The study was conducted by scientists at the University of Pretoria (UP) in 2019 and 2020 into the prevalence of Listeria monocytogenes in beef and beef products at abattoirs and retailers in the Gauteng, Mpumalanga and North West provinces.
It showed that 4.6 percent of chilled carcasses sampled at seven abattoirs in Gauteng were positive for Listeria. This means that contaminated items could enter the food chain as beef products sold at retail outlets in the province.
The study, funded by Red Meat Research and Development South Africa, was prompted by the 2017-2018 outbreak of listeriosis with 1,065 confirmed cases and 218 deaths. It was traced to a ready-to-eat processed meat product called polony, made at a plant in Polokwane run by Enterprise Foods, which at that time was owned by Tiger Brands.
Ready-to-eat (RTE) food, including polony, were also positive for Listeria in the current study.
FAO and WHO plan meeting on foodborne viruses
The UN Food and Agriculture Organization (FAO) and World Health Organization (WHO) are set to hold an expert meeting on viruses in food later this month.
The Joint FAO/WHO Expert Meeting on Microbiological Risk Assessment (JEMRA) event, at FAO headquarters in Rome on Sept. 18 to 22, will work on food attribution, analytical methods, and indicators of viruses in foods.
United States-based experts proposed for the meeting are Donald Schaffner, of Rutgers University; Xiang-Jin Meng, at Virginia Tech; Kali Kniel, from the University of Delaware; Lee-Ann Jaykus, at North Carolina State University; and Jacquelina Williams-Woods of the FDA.
In 2022, the Codex Committee on Food Hygiene (CCFH) asked JEMRA to provide scientific advice to inform a review of guidelines established in 2012. This was due to emerging issues associated with foodborne viruses and scientific developments.
Posted in Food Micro Blog, Food Microbiology, Food Microbiology Blog, Food Microbiology Research, Food Microbiology Testing, Food Virus, Food Virus Death, Hepatitis A, Hepatitis E, microbial contamination, Microbiological Risk Assessment, Microbiology, Microbiology Investigations, Microbiology Risk, Norovirus, Norovirus Oysters, Virus

